The following protocol describes combined cell surface and intracellular cytoplasmic protein staining with TotalSeq-B antibodies, with or without TotalSeq-B hashtag antibodies, using the 10x Chromium Fixed RNA Profiling Reagent Kits for Singleplexed Samples with Feature Barcode technology for Protein. TotalSeq-B antibodies are not intended for use with Barcode Oligo Capture protocols for 10x Chromium Flex products. This protocol has been optimized using liquid, single-antibody TotalSeq-B conjugates, and TotalSeq-B cocktails (Universal and TBNK). This protocol has not been optimized for the staining of nuclear or phosphorylated proteins.
Read through this protocol in its entirety and associated 10x Genomics user guides (Fixation of Cells & Nuclei for Chromium Fixed RNA Profiling, and Chromium Fixed RNA Profiling Reagent Kits for Singleplexed Samples with Feature Barcode technology for Protein) prior to starting. Note: This is a two-day protocol. Following cell surface protein staining, there is an overnight fixation step required prior to staining of intracellular proteins. Please plan accordingly.
Disclaimer:
10x Genomics has not validated this protocol and will provide limited support to customers using this protocol.
Protocol Overview

Reagent and Consumable List
- Cell Staining Buffer (BioLegend, Cat. No. 420201)
- Human TruStain FcX™ (Fc Receptor Blocking Solution) (BioLegend, Cat. No. 422301)
- TruStain FcX™ PLUS (anti-mouse CD16/32) (BioLegend, Cat. No. 156603)
- Fixation Buffer (BioLegend, Cat. No. 420801)
- Intracellular Staining Permeabilization Wash Buffer (10x) (BioLegend, Cat. No. 421002)
- True-Stain Monocyte Blocker™ (BioLegend, Cat. No. 426101)
- Low Protein Binding Microcentrifuge Tubes (ThermoFisher Scientific, Cat. No. 90410 or equivalent)
- UltraPure™ DNase/RNase-Free Distilled Water (ThermoFisher Scientific, Cat. No. 10977015 or equivalent)
- Vanadyl ribonucleoside complexes solution (VRC) (MilliporeSigma, Cat. No. 94742-10ML or equivalent)
- Poly (vinylsulfonic acid, sodium salt) solution (PVSA) (MilliporeSigma, Cat. No. 278424-250ML or equivalent)
- TBS, Tris Buffered Saline, 10X Solution, pH 7.4, Molecular Biology grade (Fisher BioReagents, Cat. No. BP24711)
- 12 x 75 mm Falcon™ Round-Bottom Polystyrene Tubes (Fisher Scientific, Cat. No. 14-959-1A or equivalent)
- RNaseZap (ThermoFisher Scientific, Cat. No. AM9780 or equivalent)
Additional Suggested Reagents
- Brefeldin A (BioLegend, Cat. No. 420601) or Monensin (BioLegend, Cat. No. 420701)
- Cell Activation Cocktail (without Brefeldin A) (BioLegend, Cat. No. 423302)
- TotalSeq™-B Human Universal Cocktail, V1.0, (BioLegend, Cat No. 399904)
- TotalSeq™-B Human TBNK Cocktail, (BioLegend, Cat. No. 399902)
- TotalSeq™-B Mouse Universal Cocktail, V1.0, (BioLegend, Cat. No. 199902)
- TotalSeq™-B Mouse Myeloid Cocktail, V1.0, (BioLegend, Cat. No. 199904)
Best Practices and Important Considerations for Best Results
Surface Staining
Cell washing
- When washing cells, it is extremely important to thoroughly decant the wash buffer, and upon the addition of new wash buffer, that the cell pellet is resuspended either with pipette mixing or gentle vortexing. When decanting, pour off the wash buffer in a single firm, but not forceful motion. Following decanting, continue to hold the tube inverted and remove remaining droplets on the lip of the tube by gently dabbing with clean paper towel before returning the tubes to an upright position. This technique should be used during all cell washes.
Optimal cell staining with TotalSeq™ antibodies
- Our liquid, single-antibody TotalSeq conjugates require optimization of staining concentration (titration) to obtain the best results, as performed by the end user of the antibodies. This includes TotalSeq antibodies targeting intracellular proteins of interest. Optimization of antibody staining concentration is essential to obtain good quality data in antibody-based applications such as surface and intracellular staining using TotalSeq antibodies.
- Antibody titration for sequencing-based applications using TotalSeq antibodies is best performed using matching procedures as much as possible. This means titration-by-sequencing if possible. In the absence of titration-by-sequencing, it may be possible to replicate this protocol using fluorescent antibodies of the same clone and titrate via flow cytometry, however, this may prove challenging for TotalSeq intracellular antibodies as optimal concentration via flow cytometry may not correlate well with optimal concentration for detection of intracellular targets by sequencing. In general, we have found that most intracellular antibodies can be used at a concentration between 0.1 – 0.5 µg. BioLegend can provide recommended concentration ranges for most TotalSeq antibodies, including intracellular antibodies, and these can be obtained by contacting BioLegend Technical Services.
- Throughout this protocol the term “antibody pool” is defined as a user-created antibody pool of titrated single TotalSeq antibodies.
- BioLegend’s lyophilized Human TotalSeq-B antibody cocktails have been validated for use with this application. The cocktails are optimized for specific staining reaction sizes. The TotalSeq-B Human TBNK Cocktail is optimized to stain 1 x 106 cells in 100 µL staining volume, and the TotalSeq-B Human Universal Cocktail V1.0 has been optimized to stain 5 x 105 cells in 50 µL staining volume. Note that for the purpose of this application, we have validated staining of up to 2 x 106 cells with 1 test of the TotalSeq-B Human TBNK and Universal Cocktail with no effect on staining efficiency and can reliably detect lineage markers using this protocol. It is important that final staining volume, regardless of cell number used, not exceed 100 µL for the TBNK cocktail and 50 µL for the Universal Cocktail. For the purpose of this protocol, we do recommend staining of 1 – 2 x 106 cells to ensure sufficient cell numbers throughout the protocol.
Intracellular staining
- All solutions used for the intracellular staining section must be prepared fresh on the day of use and kept at room temperature. Prepare only the amount needed (with 10% excess) and discard any leftover solution.
- During Intracellular staining, it is essential to use proper technique to avoid ribonuclease (RNase) contamination. After fixation, perform all steps in an RNA designated area thoroughly cleaned with RNaseZap or RNase Away. Use only RNase-free consumables, and always wear gloves.
- The addition of RNase inhibitors (VRC and PVSA) to prepared buffers used in this protocol is critical. The RNase inhibitors and quantities indicated below have been optimized for use in each respective buffer detailed in this protocol. Any alteration will require further customer validation.
- The Intracellular Staining Permeabilization Wash Buffer (10X) contains non-molecular grade components; RNase presence is expected.
- The Intracellular Staining Permeabilization Wash Buffer (10x) may precipitate during storage. It is recommended to aliquot buffer and place the aliquot at 37°C prior to use. Avoid using the same aliquot for flow cytometry in order to reduce risk of further contamination with exogenous RNase.
- Vanadyl ribonucleoside complexes solution (VRC) precipitates at lower temperatures. Prior to use, it is recommended to warm up the flask to 65°C for 10 – 15 min and mix to dissolve precipitated material. Once dissolved, use RNase-free consumables to make 1 mL aliquots and store at -20°C. Aliquots will require similar heating and vortexing before use.
- The use of BioLegend Fixation Buffer DOES NOT eliminate the need for the 10x Genomics fixation step detailed in the 10x Genomics CG000478 Demonstrated Protocol Cell & Nuclei Fixation Chromium Fixed RNA Profiling. The second fixation step is essential for ADT and RNA detection and should be performed following intracellular staining as detailed at the end of this protocol.
Protocol
Cell Surface Staining
1.Prepare cell suspension with preferred or recommended method
Notes:
- This protocol has been optimized using fresh human PBMCs isolated using density gradient centrifugation. Whole blood or lysed whole blood is not recommended. If using cells isolated with a different procedure, users may need to verify the antibody staining pattern using alternative methods.
- BioLegend has not tested this protocol using single-cell suspensions derived from enzymatically digested tissue. Enzymatic digestion may result in alterations of surface protein epitopes and impact staining with TotalSeq antibodies. Optimization of staining conditions and concentrations may be required.
- Depending on your experimental set up, you may find abundant intracellular targets such as cytokines. However, it is always advisable to include a biological positive control. Activated cells can be prepared from in vivo-stimulated tissues or from in vitro-stimulated cultures (e.g. mitogen or LPS stimulation). For detection of secreted molecules such as cytokine and chemokine detection, it is critical to include a protein transport inhibitor such as Brefeldin A (BioLegend Cat. No. 420601) or Monensin (BioLegend Cat. No. 420701) in the last 4 – 6 hours of cell culture activation. For details on stimulation methods, please see our stimulation guide for cytokines/chemokines.
- Brefeldin A and Monensin will affect stability of some RNAs. Always use an unstimulated control treated with the inhibitor of choice to obtain more accurate results. If interested in a particular set of genes, it is recommended to first test inhibitor kinetics by qPCR to increase chance of transcript detection during single-cell experiments.
2. Count and assess cell viability
2.1 Using your preferred method, carefully count all cells to ensure accurate quantitation and assess cell viability.
Note:
Contact BioLegend Technical Services with any questions regarding cell viability. Ideal cell viability is >95%. Low cell viability is associated with poor single-cell sequencing data. If low cell viability is observed, users may need to enrich live cells or repeat cell suspension preparation.
3. Dilute cells in an appropriate volume prior to staining
3.1 When using an antibody pool or the TotalSeq-B TBNK Cocktail:
3.1.1 If working with human cells, dilute 1 – 2 x 106 cells in 45 μL of Cell Staining Buffer in a 12 x 75 mm flow cytometry tube.
3.1.2 If working with mouse cells, dilute 1 – 2 x 106 cells in 49.5 μL of Cell Staining Buffer in a 12 x 75 mm flow cytometry tube.
3.2 When using a TotalSeq-B Universal Cocktail:
3.2.1 If working with human cells, dilute 1 – 2 x 106 cells in 22.5 μL of Cell Staining Buffer in a 12 x 75mm flow cytometry tube.
3.2.2 If working with mouse cells, dilute 1 – 2 x 106 cells in 24.75 μL of Cell Staining Buffer in a 12 x 75 mm flow cytometry tube.
4. Fc receptor blocking
4.1 If using an antibody pool or the TotalSeq-B TBNK Cocktail:
4.1.1 For human cells, add 5 μL of Human TruStain FcX™ to 1 – 2 x 106 cells in 45 µL of Cell Staining Buffer (total volume = 50 μL).
4.1.2 For mouse cells, add 0.5 µL of TruStain FcX™ PLUS (anti-mouse CD16/32) to 1 – 2 x 106 cells in 49.5 µL of Cell Staining Buffer (total volume = 50 μL).
4.2 If using the TotalSeq™-B Universal Cocktail:
4.2.1 For human cells, add 2.5 μL of Human TruStain FcX™ Fc blocking reagent to 1 – 2 x 106 cells in 22.5 µL of Cell Staining Buffer (total volume = 25 μL).
4.2.2 For mouse cells, add 0.25 μL of TruStain FcX™ PLUS (anti-mouse CD16/32) to 1 – 2 x 106 cells in 24.75 µL of Cell Staining Buffer (total volume = 25 μL).
4.3 Incubate for 10 minutes at 4°C.
4.4 While cells are incubating in Fc Block, proceed to step 5 – Stain cells with cell surface antibodies.
5. Stain cells with cell surface antibodies
Notes:
If you plan to multiplex samples using TotalSeq hashtags, there are two commonly used approaches to staining samples with hashtag antibodies.
- In the first approach, individual samples are stained with TotalSeq antibodies (user-created antibody pool or with our pre-optimized TotalSeq-B cocktails) and hashtags in a single step, washed three times to remove unbound antibodies, and subsequently pooled for loading into one or more lanes on the 10x Genomics instrument. See figure 1A.
- The second approach involves first staining individual samples with hashtags, washing three times, sample pooling, and then staining with the TotalSeq antibodies (user-created antibody pool or with our pre-optimized TotalSeq-B cocktails), followed by another three washes. See figure 1B.
- Whenever possible, we recommend the first approach above as the second approach can result in increased sample loss and reduced cell viability. Reducing sample washing will result in higher background and poor data quality. Additional information regarding use of TotalSeq hashtag antibodies can be found here – Efficient Multiplexing With TotalSeq Hashtags.
- For assistance in de-multiplexing after sequencing, please contact BioLegend Technical Services.
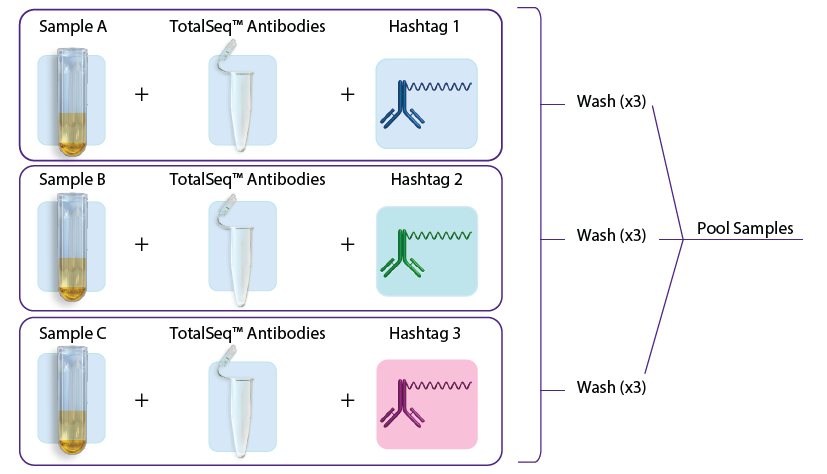
A.
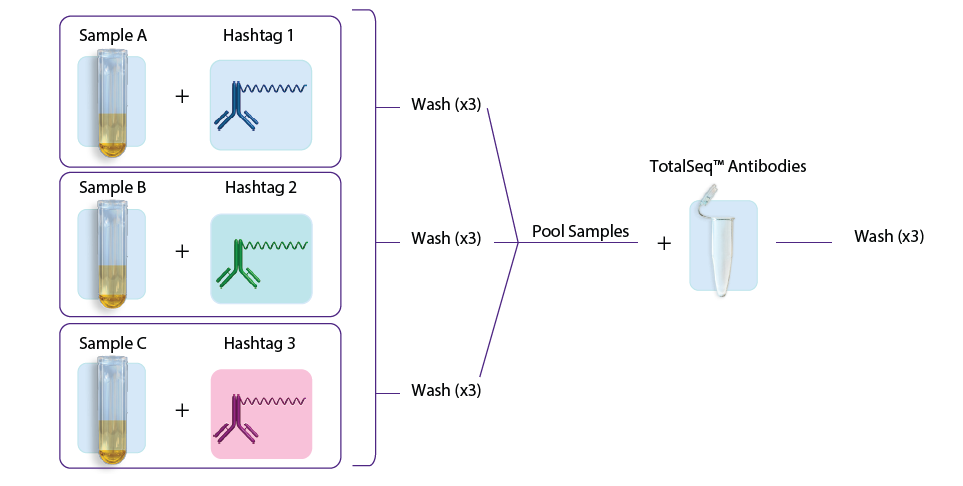
B.
Figure 1: A. Staining with TotalSeq antibodies and hashtags in a single step before pooling samples. B. Staining individual samples with hashtag reagents followed by pooling and staining with additional TotalSeq antibodies in subsequent steps.
5.1 Staining individual samples with TotalSeq™ Hashtag Antibodies for sample pooling
Notes:
- If you are not using TotalSeq Hashtags, or if you are staining cells simultaneously with an antibody pool or one of our TotalSeq-B cocktails and hashtags in a single step before pooling samples (figure 1A), proceed to step 5.2.
- If sequentially staining individual samples with hashtag antibodies, pooling samples, then staining with a TotalSeq-B antibody pool or one of our TotalSeq B cocktails as seen in figure 1B, follow the steps below before staining your samples with an antibody pool or cocktail. Please note that when sequentially staining, cell recovery may be diminished and cell viability may be negatively impacted in some cases.
5.1.1 Make a unique hashtag staining solution for each sample using a titrated amount of each hashtag in Cell Staining Buffer. Add the calculated amount of each hashtag antibody to a low protein binding microcentrifuge tube, and bring up the total volume with Cell Staining buffer to 50 µL if using an antibody pool or TotalSeq-B TBNK cocktail. Or, bring the total volume up to 25 µL if using the TotalSeq-B Universal Cocktail.
5.1.2 Centrifuge the hashtag solution at 14,000 x g at 2 – 8°C for 10 minutes before adding to the cells to ensure removal of protein aggregates.
5.1.3 Carefully pipette out the prepared hashtag solution, avoiding the bottom of the tube, and add it to the 50 µL or 25 µL blocked cell suspension.
5.1.4 Incubate for 30 minutes at 4°C.
5.1.5 Wash cells, add 3 mL of Cell Staining Buffer, mix by gently pipetting 5 times. Centrifuge at 4°C for 5 minutes at 400 – 600 x g depending on your sample type.
5.1.6 Repeat wash twice for a total of 3 washes.
Note:
It is extremely important to thoroughly decant the wash buffer and resuspend the cell pellet either with pipetting or gentle vortexing. Discard supernatant with a single firm, but not forceful motion. Proceed to absorb any remaining liquid on the lip of the tube with a clean paper towel.
5.1.7 After the final wash, resuspend each sample in 100 µL of Cell Staining Buffer, and verify cell concentration for each sample.
5.1.8 Based on each sample’s cell concentration, combine an equal number of cells from each sample to achieve 1 – 2 x 106 cells total in a low protein binding microcentrifuge tube. The final volume will be dependent on the specific staining reaction size of the antibody pool or cocktail being used.
If staining cells with an antibody pool or the TotalSeq-B TBNK cocktail and the volume of combined cells is less than 50 µL, adjust volume with Cell Staining Buffer up to 50 µL to achieve 1 – 2 x 106 cells/50 µL. If the final volume of combined cells is greater than 50 µL, centrifuge at 4°C for 5 minutes at 400 – 600 x g. Following centrifugation, adjust volume to 50 µL by removing Cell Staining Buffer. After addition or removal of Cell Staining Buffer, gently resuspend cells by pipetting.
If staining cells with the TotalSeq-B Universal cocktail and the volume of combined cells is less than 25 µL, adjust volume with Cell Staining Buffer up to 25 µL to achieve 1 – 2x 106 cells/25 µL. If the final volume of combined cells is greater than 25 µL, centrifuge at 4°C for 5 minutes at 400 – 600 x g. Following centrifugation, adjust volume to 25 µL by removing Cell Staining Buffer. After addition or removal of Cell Staining Buffer, gently resuspend cells by pipetting.
Examples:
- If you combined 6 samples at 10 µL each for a total of 60 µL and used an antibody pool or TotalSeq-B TBNK Cocktail, you would remove 10 µL after centrifugation for a total volume of 50 µL, resulting in 1 – 2 x 106 cells/50 µL.
- If you combined 4 samples at 10 µL each for a total of 40 µL and used the TotalSeq-B Universal Cocktail to stain cells, you would remove 15 µL of buffer after centrifugation for a total of 25 µL, resulting in 1 – 2 x 106 cells/25 µL.
5.1.9 Proceed with staining your combined samples using either the Cell Staining with TotalSeq with Antibody Pool section 5.2.1, or Cell Staining with TotalSeq-B Universal Cocktail or TotalSeq-B TBNK Cocktail section protocols below. Cells will already be stained with TotalSeq hashtag antibodies; do not add additional hashtag antibodies to the TotalSeq-B antigen specific antibody pool.
5.2 Cell Staining with TotalSeq™ Antibody Pool or Cocktails and TotalSeq™ Hashtag Antibodies
5.2.1 Cell Staining with TotalSeq™ Antibody Pool and TotalSeq™ Hashtags
5.2.1.1 Prepare antibody pool by combining titrated amounts of each specific TotalSeq-B antibody in a low protein binding microcentrifuge tube. For more information regarding titration of TotalSeq-B antibodies, please read Tips and Tricks for Titrating TotalSeq Antibodies. If you have additional questions, please reach out to BioLegend Technical Services.
5.2.1.2 When performing dual staining with TotalSeq-B cell hashtag antibodies and TotalSeq-B antigen specific antibodies, we recommend adding cell hashing antibodies into each respective sample’s TotalSeq-B antibody pool (figure 1A).
5.2.1.3 If the antibody pool volume is less than 50 µL, adjust volume with Cell Staining Buffer up to 50 µL. If the volume of the pool is above 50 µL, no volume adjustment is necessary.
5.2.1.4 Centrifuge the antibody pool at 14,000 x g at 2 – 8°C for 10 minutes before adding to the cells. This is critical to ensure removal of protein aggregates.
5.2.1.5 Carefully pipette out the prepared antibody pool, avoiding the bottom of the tube, and add the TotalSeq antibody pool to the 50 µL blocked cell suspension.
5.2.1.6 Incubate for 30 minutes at 4°C.
5.2.2 Cell Staining with TotalSeq™-B Universal or TotalSeq™-B TBNK Cocktail
5.2.2.1 If staining cells with a lyophilized TotalSeq-B Universal or TBNK Cocktail, follow the reconstitution steps below.
5.2.2.2 Equilibrate the lyophilized panel vial(s) to room temperature for 5 minutes.
5.2.2.3 Place lyophilized panel vial in an empty Eppendorf tube. Spin down at 10,000 x g for 30 seconds at room temperature.
5.2.2.4 If cell hashing samples simultaneously with the TotalSeq-B Universal, or TBNK Cocktail, please refer to the “Spike-In” Guidance protocol prior to reconstitution of the lyophilized cocktail to determine how to make the Cell Staining Buffer + spike-in reconstitution mix. Then proceed to the next step, 5.2.2.5, to reconstitute the lyophilized cocktail. If not hashing or if your samples are already stained with hashtags, proceed to the next step, 5.2.2.5.
5.2.2.5 If using the TotalSeq-B Universal Cocktail, rehydrate by adding 27.5 µL of Cell Staining Buffer or the Cell Staining Buffer + hashtag mix. If using the TBNK cocktail, rehydrate by adding 50 µL of Cell Staining Buffer or the Cell Staining Buffer + hashtag mix. Replace the cap and vortex for 10 seconds.
5.2.2.6 Incubate at room temperature for 5 minutes.
5.2.2.7 Vortex again and spin down at 10,000 x g for 30 seconds at room temperature.
5.2.2.8 Transfer the entire volume, 27.5 µL for the TotalSeq-B Universal Cocktail, or 50 µL for the TotalSeq-B TBNK Cocktail, to a low protein binding Eppendorf tube.
5.2.2.9 Centrifuge at 14,000 x g for 10 min at 4°C. Important: Centrifugation of the reconstituted cocktail at 14,000 x g at 2 – 8°C for 10 minutes is critical to ensure removal of antibody aggregates.
5.2.2.10 Transfer 25 µL or 50 µL of reconstituted cocktail you used to the tube containing 25 µL or 50 µL of FcR-blocked cells, taking care to avoid the bottom of the tube. The final staining volume will be 50 µL for cells stained with the TotalSeq-B Universal Cocktail, or 100 µL for cells stained with the TotalSeq-B TBNK Cocktail. Mix by gently pipetting 5 times.
5.2.2.11 Incubate for 30 minutes at 4°C.
6. Wash cells
6.1 Add 3 mL of Cell Staining Buffer and mix by gently pipetting 5 times. Centrifuge at 4°C for 5 minutes at 400 – 600 x g depending on your sample type. Repeat step twice for a total of 3 washes.
Note:
It is important to thoroughly decant the wash buffer and resuspend the cell pellet either with pipetting or gentle vortexing. Discard supernatant with a single firm, but not overly forceful motion. Proceed to absorb any remaining liquid on the lip of the tube with a clean paper towel.
6.2 After the final wash, decant the supernatant and gently mix the cells by pulse vortexing the cell pellet at medium intensity in the residual volume. Residual volume should be between 50 – 100 µL.
6.3 Proceed immediately to step 7 – Fixation.
7. Fixation
7.1 Using the same polystyrene tube, add 1 mL of Fixation Buffer to cells from previous step. Pipette and mix 10 times with pipette set to 1 mL.
7.2 Fix cells overnight at 4°C.
7.2.1 Optionally fix for 20 minutes at room temperature, however, best RNA quality is obtained with overnight fixation.
7.3 After fixation proceed to Intracellular Staining.
Intracellular Staining
Prepare the solutions below before initiating step 8. VRC should be added warm to each solution.
- 1X TBS
| Reagent | Stock concentration | Final Concentration | Volume (1 reaction + 10%) |
| TBS | 10X | 1X | 330 μL |
| VRC | 200 mM | 2 mM | 33 μL |
| PVSA | 30% | 0.1% | 11 μL |
| Water | - | - | 2.926 mL |
- 1X Intracellular Staining Permeabilization Wash Buffer
| Reagent | Stock concentration | Final Concentration | Volume (1 reaction + 10%) |
| Intracellular Staining Permeabilization Wash Buffer | 10X | 1X | 1.4 mL |
| VRC | 200 mM | 5 mM | 350 μL |
| PVSA | 30% | 0.1% | 50 μL |
| Water | - | - | 12.2 mL |
- Blocking Cocktail
| Reagent | Stock concentration | Final Concentration | Volume (1 reaction + 10%) |
| Intracellular Staining Permeabilization Wash Buffer | 10X | 1X | 2.75 μL |
| VRC | 200 mM | 30 mM | 4.12 μL |
| PVSA | 30% | 3% | 2.75 μL |
| TruStain FcX™ Block | 1X | 1X | 5.5 μL |
| True-Stain Monocyte Blocker | 1X | 1X | 5.5 μL |
| Water | - | - | 4.38 µL |
- Intracellular Antibody Pool
TotalSeq™-B Intracellular Antibody Pool Preparation
- In a new, clean, low protein binding microcentrifuge tube, prepare the intracellular antibody pool using titrated amounts of each TotalSeq-B intracellular antibody for each sample. We have found that most TotalSeq intracellular antibodies can be used at a concentration between 0.1 – 0.5 μg. For more information regarding TotalSeq antibody concentrations, please reach out to BioLegend Technical Services.
- After antibodies have been combined, bring the volume up to 48 µL with UltraPure™ DNase/RNase-Free Distilled Water. DO NOT dilute the antibodies with Intracellular Staining Permeabilization Buffer in this step, UltraPure™ DNase/RNase-Free Distilled Water must be used. Dilution with Intracellular Staining Permeabilization Buffer during this step can contribute to increased aggregate formation.
- Centrifuge the antibody pool at 14,000 x g at 2 – 8°C for 10 minutes. This is critical to ensure removal of protein aggregates.
- Following centrifugation, carefully pipette out 45 µL of the prepared antibody pool, avoiding the bottom of the tube, and add the TotalSeq antibody pool to a new, clean, low protein binding microcentrifuge tube.
- Proceed with adding 10x Intracellular Staining Permeabilization Wash Buffer as described in the table below to the antibody pool and mix by pipetting 5 times.
- The Antibody Pool can be stored at room temperature until ready for use in step 10 below – Stain cells with intracellular antibodies.
| Reagent | Stock concentration | Final Concentration | Volume (1 reaction) |
| Intracellular Staining Permeabilization Wash Buffer | 10X | 1X | 5 μL |
| Antibody pool + Water | 1X | 1X | 45 μL |
8. Quenching
8.1 Quench fixation by adding 100 μL 10X TBS to each tube and gently vortex. Incubate tubes at room temperature for 5 minutes.
8.2 Add 3 mL of prepared 1X TBS to each tube and centrifuge tubes at 850 x g at room temperature for 5 minutes; discard supernatant.
9. Permeabilization and blocking
9.1 Resuspend fixed cells with 1 mL of prepared 1X Intracellular Staining Permeabilization Wash Buffer. Mix by gently pipetting 5 times.
9.2 Following resuspension, add an additional 1 mL of prepared 1X Intracellular Staining Permeabilization Wash Buffer to the resuspended cells. Centrifuge at 850 x g at room temperature for 5 minutes.
9.3 Repeat step for a total of 2 washes.
9.4 After the final wash, decant the supernatant and gently mix the cells by pipetting the residual volume 5 times. Residual volume should be between 50 – 100 µL.
9.5 Add 25 µL of prepared Blocking Cocktail to the residual volume of cells.
Note:
True-Stain Monocyte Blocker™ is used to reduce non-specific background of intracellular TotalSeq conjugates.
9.6 Gently mix the cells by pipetting 5 times and incubate for 15 minutes at room temperature.
10. Stain cells with intracellular antibodies
10.1 Add 50 µL of the prepared Intracellular Antibody Cocktail to blocked cells. Mix by pipetting 5 times and stain for 30 - 60 minutes at room temperature.
10.2 Add 1 mL of prepared 1x Intracellular Staining Perm Wash Buffer. Mix by gently pipetting 5 times. Following resuspension, add an additional 2 mL of prepared 1x Intracellular Staining Perm Wash Buffer. Centrifuge at room temperature for 5 minutes at 850 x g. Decant supernatant and repeat wash 2 more times for a total of 3 washes.
10.3 Following the final wash and after decanting supernatant, resuspend the cells in residual volume by pipetting cell pellet 5 times. Residual volume should be between 50 – 100 µL.
10.4 Transfer resuspended cells to a new low protein binding microcentrifuge tube.
10.5 Wash the polystyrene tube with 100 µL 1x Intracellular Staining Perm Wash Buffer. Combine wash liquid with resuspended cells.
10.6 Centrifuge tube at room temperature for 5 minutes at 850 x g. Carefully remove all supernatant. Proceed immediately to step C in the Sample Fixation section of the 10x CG000478 Demonstrated Protocol Cell & Nuclei Fixation Chromium Fixed RNA Profiling.
Recommended sequencing depth for cell surface protein library
To obtain sufficient read coverage for Cell Surface Protein libraries, follow recommended library loading and pooling specifications provided in 10x Genomics user guides. See table below for BioLegend sequencing depth recommendations for Cell Surface and Intracellular Protein libraries.
| Library Type | Minimum Sequencing Depth |
|---|---|
| Cell Surface and Intracellular Protein Library <100 Antibody Derived Tag (ADT) panel | 5,000 |
| Cell Surface and Intracellular Protein Library ≥100 ADT panel | 10,000 |
Disclaimer: Buyer is solely responsible for determining whether Buyer has all intellectual property rights that are necessary for Buyer's intended uses of the BioLegend TotalSeq products. For example, for any technology platform Buyer uses with TotalSeq, it is Buyer's sole responsibility to determine whether it has all necessary third-party intellectual property rights to use that platform and TotalSeq with that platform.